publications
2025
-
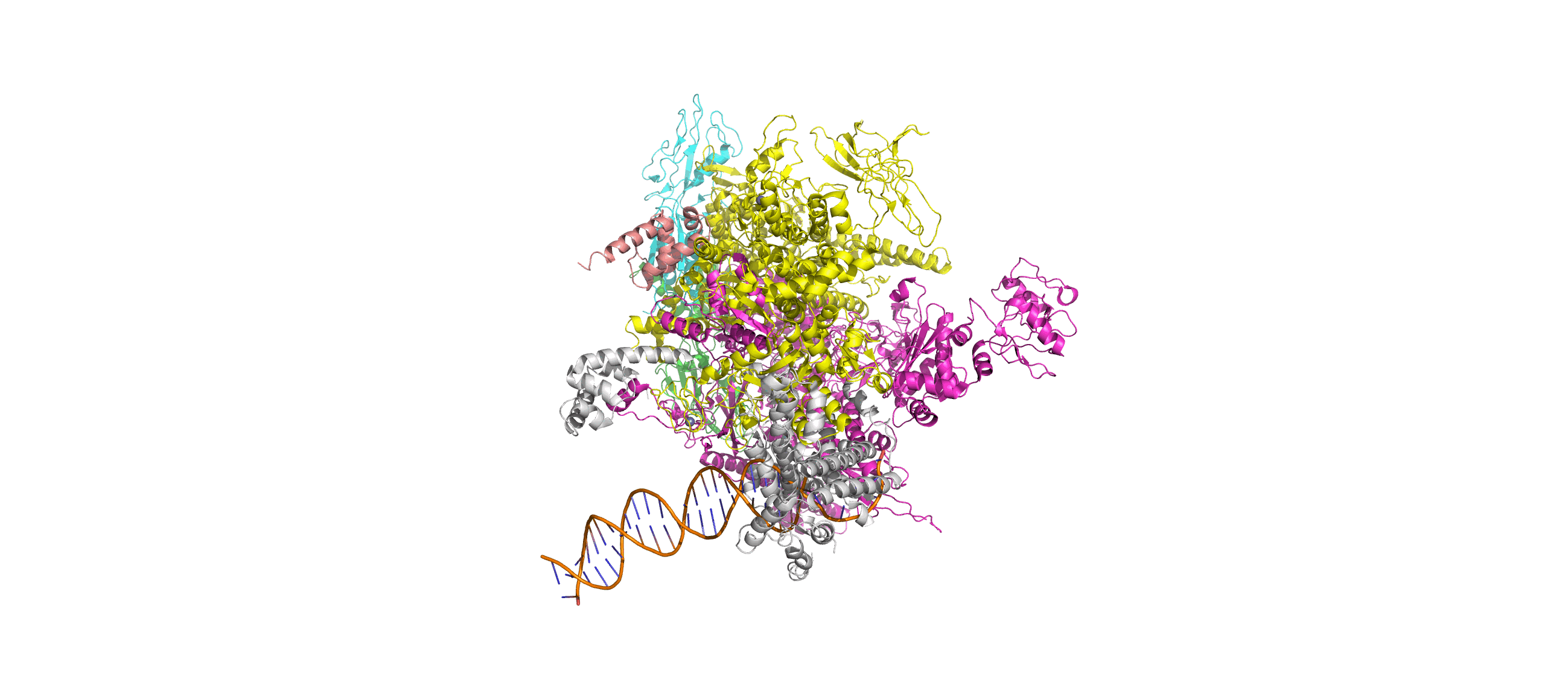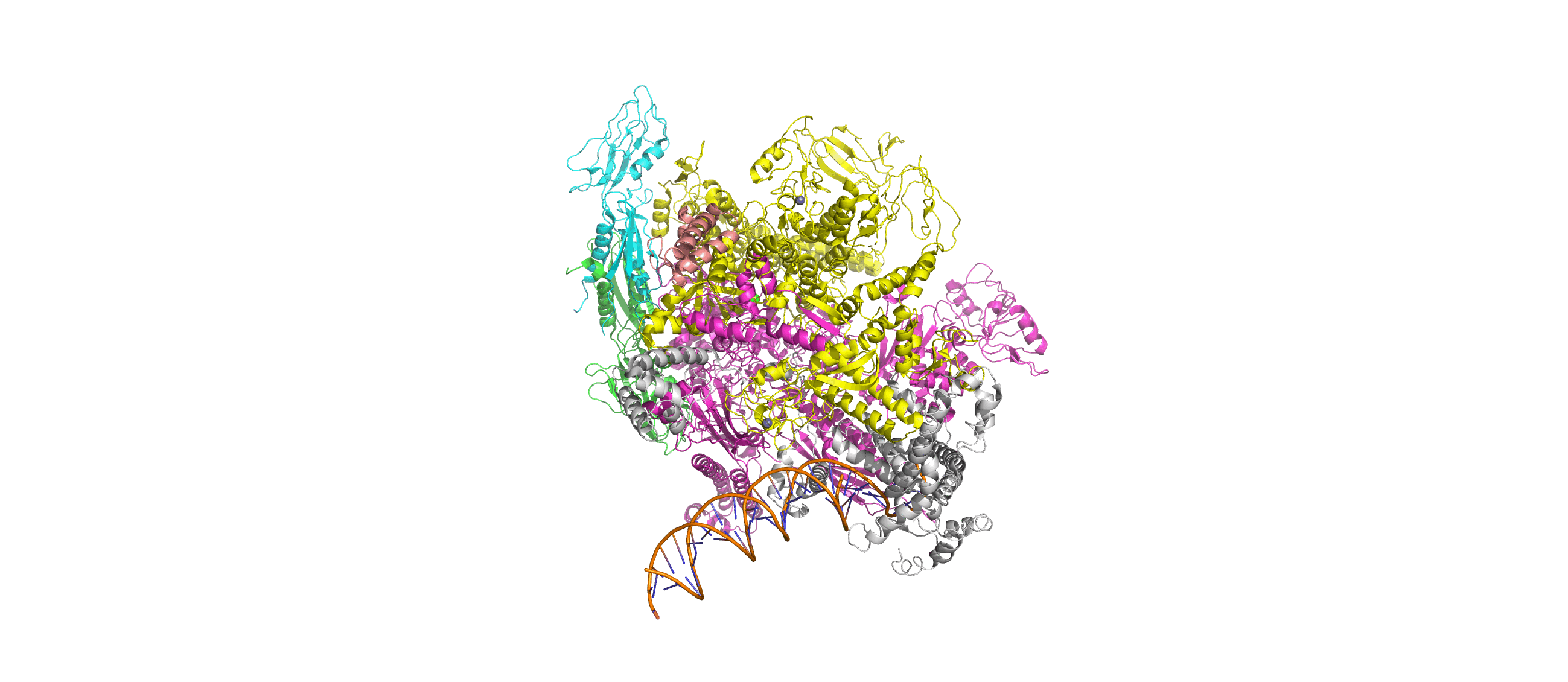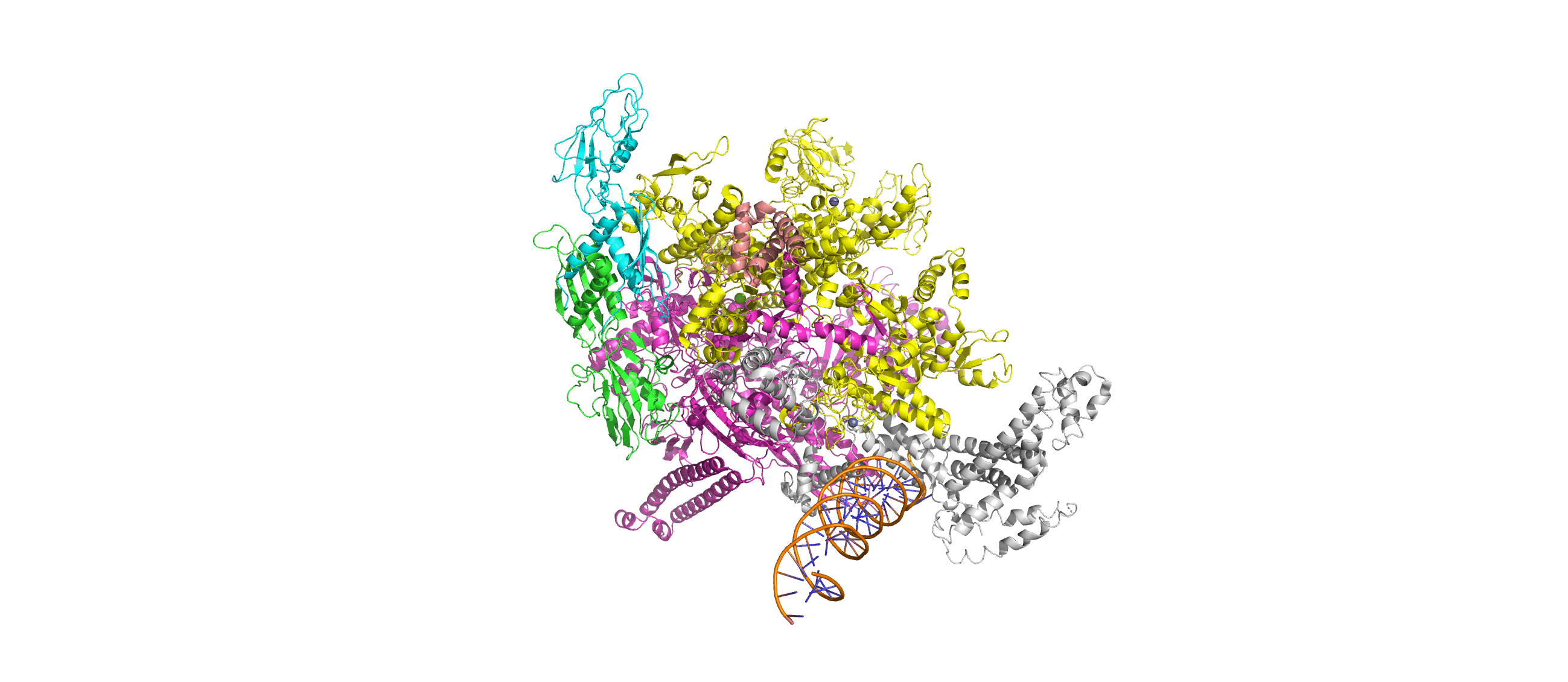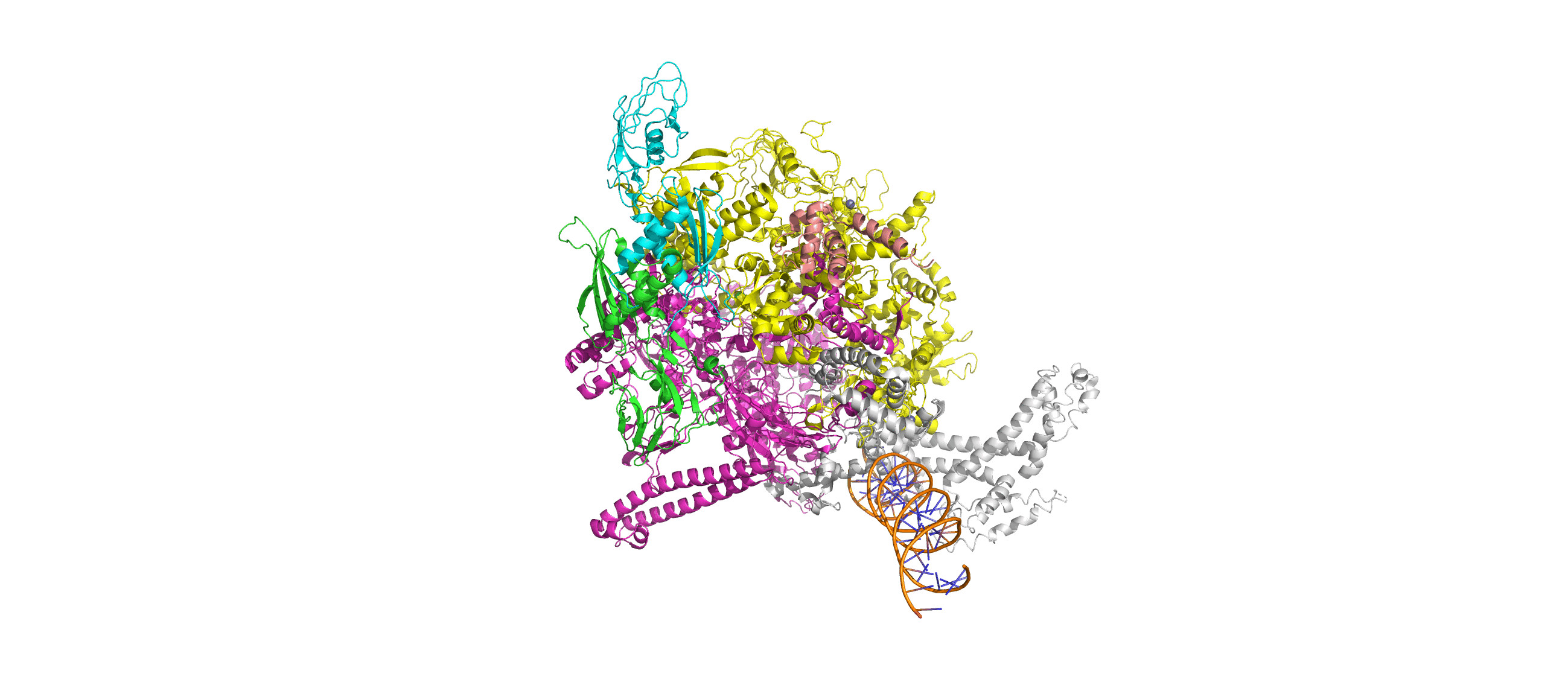 Computation-Guided Redesign of Promoter Specificity of a Bacterial RNA PolymeraseXiangyang Liu, Anthony T. Meger, Thomas G. Gillis, Jonah O’Mara Schwartz, and 3 more authorsbioRxiv, Jun 2025*Equal contribution by Liu, Meger, Gillis, and O’Mara Schwartz
Computation-Guided Redesign of Promoter Specificity of a Bacterial RNA PolymeraseXiangyang Liu, Anthony T. Meger, Thomas G. Gillis, Jonah O’Mara Schwartz, and 3 more authorsbioRxiv, Jun 2025*Equal contribution by Liu, Meger, Gillis, and O’Mara SchwartzThe ability to regulate circuits and pathways is central to cellular control. The existing toolkit is predominantly comprised of local transcription regulators that are unsuitable for exerting control at a global genome-wide scale. Bacterial sigma factors are ideal global regulators as they direct the RNA polymerase to thousands of transcription sites. Here, we redesigned the promoter specificity of the E. coli housekeeping sigma factor, sigma-70, toward five orthogonal promoter targets not recognized by the native sigma-70. These orthogonal sigma-70s were engineered by screening a pooled library of computationally-guided designs customized toward each promoter target. A combination of conserved interactions with the DNA backbone and target-specific interactions facilitate new promoter recognition. Activity of the top performing redesigned sigma-70s varied across the promoter targets and ranged from 17% to 77% of native sigma-70 on its canonical active promoter. These orthogonal sigma factors represent a new suite of regulators for global transcriptional control.
2020
-
 A Gnotobiotic System for Studying Microbiome Assembly in the Phyllosphere and in Vegetable FermentationEsther R. Miller, Jonah O’Mara Schwartz, Grace Cox, and Benjamin E. WolfeJoVE, Jun 2020Publisher: MyJoVE Corp
A Gnotobiotic System for Studying Microbiome Assembly in the Phyllosphere and in Vegetable FermentationEsther R. Miller, Jonah O’Mara Schwartz, Grace Cox, and Benjamin E. WolfeJoVE, Jun 2020Publisher: MyJoVE CorpThe phyllosphere, the above ground portion of the plant that can be colonized by microbes, is a useful model system to identify processes of microbial community assembly. This protocol outlines a system for studying microbial community dynamics in the phyllosphere of Napa cabbage plants. It describes how to grow germ-free plants in test tubes with a calcined clay and nutrient broth substrate. Inoculation of germ-free plants with specific microbial cultures provides opportunities to measure microbial growth and community dynamics in the phyllosphere. Through the use of sterile vegetable extract produced from cabbages shifts in microbial communities that occur during fermentation can also be assessed. This system is relatively simple and inexpensive to set up in the lab and can be used to address key ecological questions in microbial community assembly. It also provides opportunities to understand how phyllosphere community composition can impact the microbial diversity and quality of vegetable fermentations. This approach for developing gnotobiotic cabbage phyllosphere communities could be applied to other wild and agricultural plant species.
2019
-
 Establishment Limitation Constrains the Abundance of Lactic Acid Bacteria in the Napa Cabbage PhyllosphereEsther R. Miller, Patrick J. Kearns, Brittany A. Niccum, Jonah O’Mara Schwartz, and 2 more authorsApplied and Environmental Microbiology, Jun 2019Publisher: American Society for Microbiology
Establishment Limitation Constrains the Abundance of Lactic Acid Bacteria in the Napa Cabbage PhyllosphereEsther R. Miller, Patrick J. Kearns, Brittany A. Niccum, Jonah O’Mara Schwartz, and 2 more authorsApplied and Environmental Microbiology, Jun 2019Publisher: American Society for MicrobiologyPatterns of phyllosphere diversity have become increasingly clear with high-throughput sequencing surveys, but the processes that control phyllosphere diversity are still emerging. Through a combination of lab and field experiments using Napa cabbage and lactic acid bacteria (LAB), we examined how dispersal and establishment processes shape the ecological distributions of phyllosphere bacteria. We first determined the abundance and diversity of LAB on Napa cabbage grown at three sites using both culture-based approaches and 16S rRNA gene amplicon sequencing. Across all sites, LAB made up less than 0.9% of the total bacterial community abundance. To assess whether LAB were low in abundance in the Napa cabbage phyllosphere due to a limited abundance in local species pools (source limitation), we quantified LAB in leaf and soil samples across 51 vegetable farms and gardens throughout the northeastern United States. Across all sites, LAB comprised less than 3.2% of the soil bacterial communities and less than 1.6% of phyllosphere bacterial communities. To assess whether LAB are unable to grow in the phyllosphere even if they dispersed at high rates (establishment limitation), we used a gnotobiotic Napa cabbage system in the lab with experimental communities mimicking various dispersal rates of LAB. Even at high dispersal rates, LAB became rare or completely undetectable in experimental communities, suggesting that they are also establishment limited. Collectively, our data demonstrate that the low abundance of LAB in phyllosphere communities may be explained by establishment limitation.IMPORTANCE The quality and safety of vegetable fermentations are dependent on the activities of LAB naturally present in the phyllosphere. Despite their critical role in determining the success of fermentation, the processes that determine the abundance and diversity of LAB in vegetables used for fermentation are poorly characterized. Our work demonstrates that the limited ability of LAB to grow in the cabbage phyllosphere environment may constrain their abundance on cabbage leaves. These results suggest that commercial fermentation of Napa cabbage proceeds despite low and variable abundances of LAB across different growing regions. Propagule limitation may also explain ecological distributions of other rare members of phyllosphere microbes.